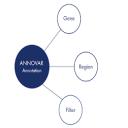
ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. ANNOVAR is an efficient software tool to utilize update-to-date information to functionally annotate genetic variants detected from diverse genomes (including human genome hg18, hg19, as well as mouse, worm, fly, yeast and many others).ANNOVAR is written in Perl and can be run as a standalone application on diverse hardware systems where standard Perl modules are installed.
...continue to readProDeGe is the first fully automated computational protocol for decontamination of genomes. The protocol uses public databases to detect contamination in agenome assembly, then groups contigs into “Clean” or “Contaminant” groups. ProDeGe spits out lists that users can read through to identify contaminants and determine what they mightbe.The standalone software is freely available and can be run on any system that has Perl, R, Prodigal and NCBI Blast installed. (Tennessen et al., ISME J.
...continue to readThe DAGchainer software computes chains of syntenic genes found within complete genome sequences. As input, DAGchainer accepts a list of gene pairs with sequence homology along with their genome coordinates. Using a scoring function which accounts for the distance between neighboring genes on each DNA molecule and the BLAST E-value score between homologs, maximally scoring chains of ordered gene pairs are computed and reported. This algorithm can be used to mine large evolutionary conserved regi
...continue to readGalaxy is an open, web-based platform for data intensive biomedical research, now available as a "cloud computing" resource Whether on this free public server or your own instance, you can perform, reproduce, and share complete analyses.
...continue to readVarSifter is a graphical Java program designed to display, sort, filter, and generally sift variation data from massively parallel sequencing experiments. It is designed to read exome-scale variation data in either a tab-delimited text file with header, or an uncompressed VCF file. The program is written in Java, and should run on any platform with a current Java Virtual Machine.
...continue to readArrayOligoSelector (AOS) systematically designs gene specific long oligo probes for entire genomes. The program optimizes the oligo selections for several parameters, including uniqueness in the genome, internal repeats, self-binding, and GC content.
...continue to readPLINK is a popular and computationally efficient software program that offers a comprehensive and well-documented set of automated genome wide association quality control and analysis tools. It is a freely available open source software written in C++, which can be installed on Windows, Mac and Unix machines. The focus of PLINK is purely on analysis of genotype/phenotype data, so there is no support for steps prior to this (e.g. study design and planning, generating genotype or CNV calls from ra
...continue to readA portal collecting several downloadable tools for the study of genomic structural & copy number variations. Software tools can be used to construct a personal diploid genome sequence by including personal variants into reference genome (vcf2diploid), to discover and genotype CNVs from genome sequencing (CNVnator), etc.
...continue to readThe R package multiMiR, with web server is a comprehensive collection of predicted and validated miRNA-target interactions and their associations with diseases and drugs. Within this database, investigators can find clues to potential new treatments for various diseases, including cancer. In addition to helping researchers search for relationships between microRNAs and their genetic targets, multiMiR includes drugs that affect these microRNAs and lists associated diseases.The database combines n
...continue to readThe Exomiser is a Java applicationthat that finds potential disease-causing variants from whole-exome or whole-genome sequencing data. It is an opensource Java software package that can be downloaded and run on a single desktop computer. It filters and prioritizes candidate genes and variants from whole-exome or wholegenome sequencing data, with a special focus on phenotypic data. Users enter a patient’s clinical features and exome into the program, and Exomiser generates a scored list of cand
...continue to readGene Set Enrichment Analysis (GSEA) is a computational method that determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biological states (e.g. phenotypes).
...continue to read
Add to my favorites
Remove from my favorites
useful and fast genetic calculator
Category: Genomics software